Microscopy Cell Segmentation Using Unet 3D 3plus¶
[Paper] [Notebook] [TF Implementation] [Torch Implementation]
UNet, which is one of deep learning networks with an encoder-decoder architecture, is widely used in medical image segmentation. Combining multi-scale features is one of important factors for accurate segmentation. UNet++ was developed as a modified Unet by designing an architecture with nested and dense skip connections. However, it does not explore sufficient information from full scales and there is still a large room for improvement.
UNET 3plus full-scale skip connections convert the inter-connection between the encoder and decoder as well as intra-connection between the decoder sub-networks. Both UNet with plain connections and UNet++ with nested and dense connections are short of exploring sufficient information from full scales, failing to explicitly learn position and boundary of an organ. To remedy the defect in UNet and UNet++, each decoder layer in UNet 3+ incorporates both smaller- and same-scale feature maps from encoder and larger-scale feature maps from decoder, which capturing fine-grained details and coarse-grained semantics in full scales.
This example of UNET 3D 3plus to modification of UNET 3plus to segmentation 3D dataset. We are showcasing UNET 3D 3plus to segment electronic microscopy 3D cell dataset.
Getting things ready¶
Lets import the necessary packages:
import tempfile
import fastestimator as fe
from fastestimator.dataset.data.em_3d import load_data
from fastestimator.op.numpyop.meta import Sometimes
from fastestimator.op.numpyop.multivariate import HorizontalFlip, VerticalFlip
from fastestimator.op.numpyop.univariate import ChannelTranspose, Minmax
from fastestimator.op.numpyop.univariate.expand_dims import ExpandDims
from fastestimator.op.tensorop import TensorOp
from fastestimator.op.tensorop.loss import CrossEntropy
from fastestimator.op.tensorop.model import ModelOp, UpdateOp
from fastestimator.op.tensorop.resize3d import Resize3D
from fastestimator.op.tensorop.argmax import Argmax
from fastestimator.trace.adapt import EarlyStopping, ReduceLROnPlateau
from fastestimator.trace.io import BestModelSaver
from fastestimator.trace.metric import Dice
from fastestimator.util import ImageDisplay, GridDisplay
Let’s define several uitlitiy functions:
import copy
import matplotlib.pyplot as plt
import numpy as np
from matplotlib import animation, rc
rc('animation', html='jshtml')
def apply_mask(image, mask, color, alpha=0.1):
"""
Apply the given mask to the image.
image: the input image(H, W, D, C)
mask: the mask to overlay(H, W, D)
color_mapping: list of color value [C]
"""
channels = image.shape[-1]
for c in range(channels):
image[:, :, :, c] = np.where(mask == 1,
image[:, :, :, c] *
(1 - alpha) + alpha * color[c],
image[:, :, :, c])
return image
def generate_mask_overlay_image(image, mask, color_mapping):
"""
Generate a image overlaying mask over it.
image: the input image(H, W, D, C)
mask: the mask to overlay(H, W, D, class)
color_mapping: a dictionary mapping input classes to colors(eg: {0: [0, 0, 0], 1: [0, 40, 255]})
The dictionary should have length "class" and each element should contain list of length "C"
image and color_mapping are expected to be in same format(eg 0-1 or 0-255)
"""
combined_image = copy.deepcopy(image)
classes = mask.shape[-1]
for i in range(classes):
combined_image = apply_mask(combined_image, mask[:,:,:,i], color_mapping[i])
return combined_image
def create_animation(images, labels):
"""
create animation combining input image and mask
image: the input image(H, W, D, 3)
labels: the image to compare with(H, W, D, 3)
"""
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
ax1.axis('off')
ax2.axis('off')
im1 = ax1.imshow(images[:, :, 0,:])
im2 = ax2.imshow(labels[:, :, 0,:])
fig.show()
im = [im1, im2]
def animate_func(i):
im[0].set_array(images[:, :, i,:])
im[1].set_array(labels[:, :, i,:])
return [im]
return animation.FuncAnimation(fig,
animate_func,
frames=images.shape[-2],
interval=images.shape[-2]*4)
color_mapping = {
0: [0, 0, 0],
1: [0.0, 0.16, 1.0],
2: [0.0, 0.83, 1.0],
3: [0.49, 1.0, 0.48],
4: [0.50, 0.0, 0.0],
5: [1.0, 0.28, 0.0],
6: [1.0, 0.90, 0.0]}
def save_gif(image, label, color_mapping, file_name):
overlay_image = generate_mask_overlay_image(image, label, color_mapping)
create_animation(image, overlay_image).save(file_name)
Next, let's set up some hyperparameters related to the task:
batch_size = 1
epochs = 40
log_steps = 20
height = 256
width = 256
depth = 24
channels = 1
num_classes = 6
filters = 64
learning_rate = 1e-3
train_steps_per_epoch = None
eval_steps_per_epoch = None
save_dir = tempfile.mkdtemp()
data_dir = None
Importing Dataset¶
Electronic Microscopy 3D cell dataset, consists of 2 3D images, one 800x800x50 and the other 800x800x24. The 800x800x50 is used as training dataset and 800x800x24 is used for validation. Instead of using the entire 800x800 images, the 800x800x50 is tiled into 256x256x24 tiles with an overlap of 128 producing around 75 training images and similarly the 800x800x24 image is tiled to produce 25 validation images.
train_data, eval_data = load_data(data_dir)
print("training dataset length is {}".format(len(train_data)))
print("evaluation dataset length is {}".format(len(eval_data)))
print("dataset sample:")
print("Image Shape: ", train_data[0]['image'].shape)
print("Label Shape: ", train_data[0]['label'].shape)
training dataset length is 75 evaluation dataset length is 25 dataset sample: Image Shape: (256, 256, 24) Label Shape: (256, 256, 24, 6)
The imageis a 256x256x24 numpy array of uint16.
The label is a 256x256x24*6 encoded(6 classes) numpy array. Semantic label files classify each image voxel into one of six classes, indexed from 0-5:
| Index | Color | Class name |
| 0 | Dark Blue | Cell |
| 1 | Cyan | Mitochondria |
| 2 | Green | Alpha granule |
| 3 | Yellow | Canalicular vessel |
| 4 | Red Dense | granule body |
| 5 | Purple | granule core |
Creating Pipeline¶
Now that both training and validation datasets are created, we use Pipeline to define the preprocessing operations:
We are using HorizontalFlip and VerticalFlip as our applied data agumentations.
ExpandDims is used to expand the last channel(256x256x24)-> (256x256x24x1).
pipeline = fe.Pipeline(
train_data=train_data,
eval_data=eval_data,
batch_size=batch_size,
ops=[
Sometimes(numpy_op=HorizontalFlip(image_in="image", mask_in="label", mode='train')),
Sometimes(numpy_op=VerticalFlip(image_in="image", mask_in="label", mode='train')),
Minmax(inputs="image", outputs="image"),
ExpandDims(inputs="image", outputs="image"),
ChannelTranspose(inputs=("image", "label"), outputs=("image", "label"), axes=(3, 0, 1, 2))
])
data = pipeline.get_results(mode='train')
image = data['image'].numpy()
label = data['label'].numpy()
print("Image shape: ", image.shape," Mask shape:", label.shape)
Image shape: (1, 1, 256, 256, 24) Mask shape: (1, 6, 256, 256, 24)
Visualizing Sample Data:¶
image = np.squeeze(image[0])
label = np.transpose(label[0], (1, 2, 3, 0))
Now lets visualize pipeline output, a single slice of image with the label overlaying on it using ImageDisplay.
ImageDisplay(image=image[:,:,0], masks=label[:,:,0], title='Ground Truth').show()
If you want you visualize the all the slices of pipeline output, please use below code to save multi slice visualization as gif.
# adding ground truth layer
input_gt = np.ones(label.shape[:-1] + (1,), dtype=label.dtype)
input_gt[np.sum(label, axis=-1)==1] = 0
label_out = np.concatenate((input_gt, label), axis=-1)
image = np.tile(np.expand_dims(image, -1), 3)
save_gif(image, label_out, color_mapping, 'training_data.gif')
Lets see a pipeline out visualization of multiple slices and overlaying labels which is generated offline.
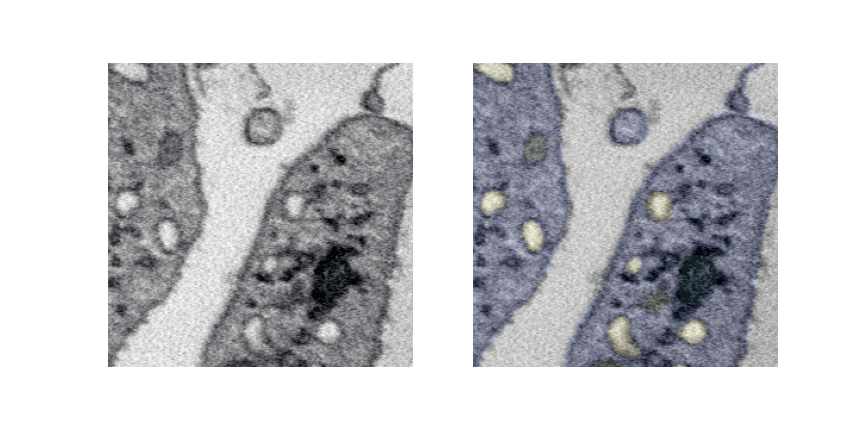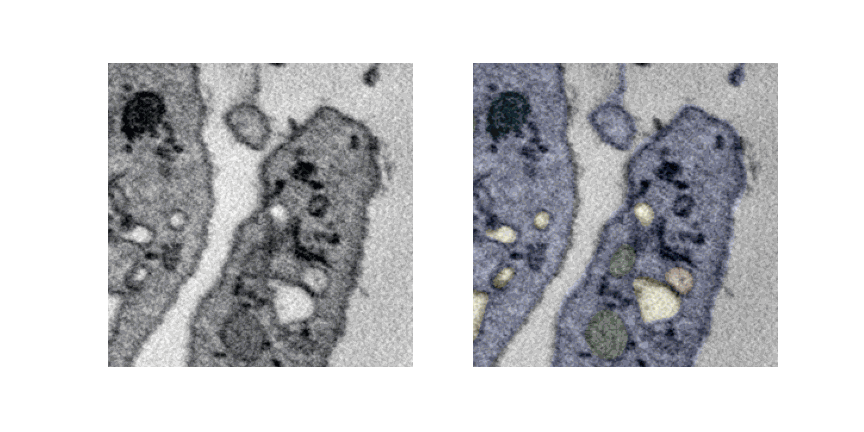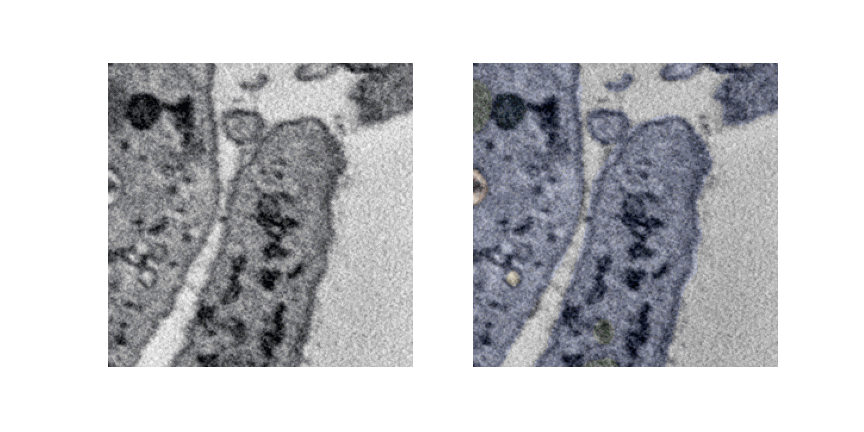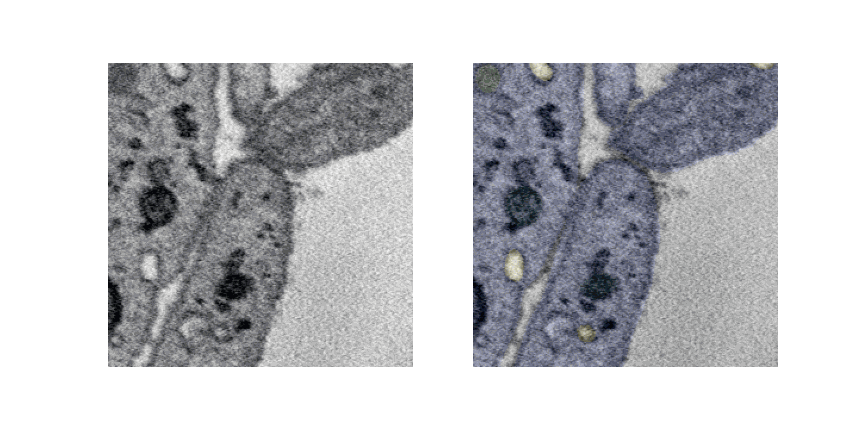
UNET 3D 3Plus¶
from typing import Tuple
import torch
import torch.nn as nn
from torch.nn import functional as F
from torch.nn.init import kaiming_normal_ as he_normal
class StdSingleConvBlock(nn.Module):
"""A UNet3D StdSingleConvBlock block.
Args:
in_channels: How many channels enter the encoder.
out_channels: How many channels leave the encoder.
"""
def __init__(self, in_channels: int, out_channels: int) -> None:
super().__init__()
self.layers = nn.Sequential(nn.BatchNorm3d(in_channels),
nn.ReLU(inplace=True),
nn.Conv3d(in_channels, out_channels, kernel_size=3, padding="same"))
for layer in self.layers:
if isinstance(layer, nn.Conv3d):
he_normal(layer.weight.data)
layer.bias.data.zero_()
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
out = self.layers(x)
return out
class ConvBlock(nn.Module):
"""A UNet3D ConvBlock block.
Args:
in_channels: How many channels enter the encoder.
out_channels: How many channels leave the encoder.
"""
def __init__(self, in_channels: int, out_channels: int) -> None:
super().__init__()
self.layers = nn.Sequential(nn.Conv3d(in_channels, out_channels, kernel_size=3, padding="same"))
for layer in self.layers:
if isinstance(layer, nn.Conv3d):
he_normal(layer.weight.data)
layer.bias.data.zero_()
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
out = self.layers(x)
return out
class StdDoubleConvBlock(nn.Module):
"""A UNet3D StdDoubleConvBlock block.
Args:
in_channels: How many channels enter the encoder.
out_channels: How many channels leave the encoder.
"""
def __init__(self, in_channels: int, out_channels: int) -> None:
super().__init__()
self.layers = nn.Sequential(
StdSingleConvBlock(in_channels, out_channels),
StdSingleConvBlock(out_channels, out_channels),
)
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
out = self.layers(x)
return out
class StdConvBlockSkip(nn.Module):
"""A UNet3D StdConvBlockSkip block skipping batch normalization.
Args:
in_channels: How many channels enter the encoder.
out_channels: How many channels leave the encoder.
"""
def __init__(self, in_channels: int, out_channels: int) -> None:
super().__init__()
self.layers = nn.Sequential(ConvBlock(in_channels, out_channels),
StdSingleConvBlock(out_channels, out_channels))
def forward(self, x: torch.Tensor) -> torch.Tensor:
out = self.layers(x)
return out
class UpsampleBlock(nn.Module):
"""A UNet3D UpsampleBlock block.
Args:
in_channels: How many channels enter the encoder.
out_channels: How many channels leave the encoder.
scale_factor: scale factor to up sample
kernel_size: size of the kernel
"""
def __init__(self, in_channels: int, out_channels: int, scale_factor: int, kernel_size: int = 3) -> None:
super().__init__()
self.layers = nn.Sequential(
nn.Upsample(scale_factor=scale_factor, mode='trilinear', align_corners=False),
nn.Conv3d(in_channels, out_channels, kernel_size, padding="same"),
)
for layer in self.layers:
if isinstance(layer, nn.Conv3d):
he_normal(layer.weight.data)
layer.bias.data.zero_()
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
out = self.layers(x)
return out
class DownSampleBlock(nn.Module):
"""A UNet3D DownSampleBlock block.
Args:
in_channels: How many channels enter the encoder.
out_channels: How many channels leave the encoder.
scale_factor: scale factor to down sample
kernel_size: size of the kernel
"""
def __init__(self, in_channels: int, out_channels: int, scale_factor: int, kernel_size: int = 3) -> None:
super().__init__()
self.scale_factor = scale_factor
self.layers = nn.Sequential(nn.Conv3d(in_channels, out_channels, kernel_size=kernel_size, padding="same"))
for layer in self.layers:
if isinstance(layer, nn.Conv3d):
he_normal(layer.weight.data)
layer.bias.data.zero_()
def forward(self, x: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:
out = self.layers(F.max_pool3d(x, self.scale_factor))
return out
class UNet3D3Plus(nn.Module):
"""A Attention UNet3D 3plus implementation in PyTorch.
Args:
input_size: The size of the input tensor (channels, height, width).
output_channel: The number of output channels.
Raises:
ValueError: Length of `input_size` is not 3.
ValueError: `input_size`[1] or `input_size`[2] is not a multiple of 16.
"""
def __init__(self,
input_size: Tuple[int, int, int] = (1, 128, 128, 24),
output_channel: int = 1,
channels: int = 64) -> None:
UNet3D3Plus._check_input_size(input_size)
super().__init__()
self.input_size = input_size
self.enc1 = StdConvBlockSkip(in_channels=input_size[0], out_channels=channels)
self.enc2 = StdDoubleConvBlock(in_channels=channels, out_channels=channels * 2)
self.enc3 = StdDoubleConvBlock(in_channels=channels * 2, out_channels=channels * 4)
self.bottle_neck = StdDoubleConvBlock(in_channels=channels * 4, out_channels=channels * 8)
self.up5_4 = UpsampleBlock(in_channels=channels * 8, out_channels=channels, scale_factor=2)
self.up5_3 = ConvBlock(in_channels=channels * 4, out_channels=channels)
self.down5_2 = DownSampleBlock(in_channels=channels * 2, out_channels=channels, scale_factor=2)
self.down5_3 = DownSampleBlock(in_channels=channels, out_channels=channels, scale_factor=4)
self.conv5 = StdSingleConvBlock(in_channels=channels * 4, out_channels=4 * channels)
self.up6_4 = UpsampleBlock(in_channels=channels * 8, out_channels=channels, scale_factor=4)
self.up6_3 = UpsampleBlock(in_channels=channels * 4, out_channels=channels, scale_factor=2)
self.up6_2 = ConvBlock(in_channels=channels * 2, out_channels=channels)
self.down6_1 = DownSampleBlock(in_channels=channels, out_channels=channels, scale_factor=2)
self.conv6 = StdSingleConvBlock(in_channels=channels * 4, out_channels=4 * channels)
self.up7_4 = UpsampleBlock(in_channels=channels * 8, out_channels=channels, scale_factor=8)
self.up7_3 = UpsampleBlock(in_channels=channels * 4, out_channels=channels, scale_factor=4)
self.up7_2 = UpsampleBlock(in_channels=channels * 4, out_channels=channels, scale_factor=2)
self.conv7_1 = ConvBlock(in_channels=channels, out_channels=channels)
self.conv7 = StdSingleConvBlock(in_channels=channels * 4, out_channels=4 * channels)
self.dec1 = nn.Sequential(nn.BatchNorm3d(channels * 4),
nn.ReLU(inplace=True),
nn.Conv3d(channels * 4, output_channel, 1, padding="same"),
nn.Sigmoid())
for layer in self.dec1:
if isinstance(layer, nn.Conv3d):
he_normal(layer.weight.data)
layer.bias.data.zero_()
def forward(self, x: torch.Tensor) -> torch.Tensor:
conv1 = self.enc1(x)
pool1 = F.max_pool3d(conv1, 2)
conv2 = self.enc2(pool1)
pool2 = F.max_pool3d(conv2, 2)
conv3 = self.enc3(pool2)
pool3 = F.max_pool3d(conv3, 2)
conv4 = self.bottle_neck(pool3)
up5_4 = self.up5_4(conv4)
up5_3 = self.up5_3(conv3)
down5_2 = self.down5_2(conv2)
down5_3 = self.down5_3(conv1)
conv5 = self.conv5(torch.cat((up5_4, up5_3, down5_2, down5_3), 1))
up6_4 = self.up6_4(conv4)
up6_3 = self.up6_3(conv5)
up6_2 = self.up6_2(conv2)
down6_1 = self.down6_1(conv1)
conv6 = self.conv6(torch.cat((up6_4, up6_3, up6_2, down6_1), 1))
up7_4 = self.up7_4(conv4)
up7_3 = self.up7_3(conv5)
up7_2 = self.up7_2(conv6)
conv7_1 = self.conv7_1(conv1)
x_out = self.dec1(self.conv7(torch.cat((up7_4, up7_3, up7_2, conv7_1), 1)))
return x_out
@staticmethod
def _check_input_size(input_size):
if len(input_size) != 4:
raise ValueError("Length of `input_size` is not 4 (channel, height, width, depth)")
_, height, width, depth = input_size
if height < 8 or not (height / 8.0).is_integer() or width < 8 or not (
width / 8.0).is_integer() or depth < 8 or not (depth / 8.0).is_integer():
raise ValueError(
"All three height, width and depth of input_size need to be multiples of 8 (8, 16, 32, 48...)")
Network operations during training¶
input_shape = (height, width, depth)
model = fe.build(model_fn=lambda: UNet3D3Plus((channels, ) + input_shape, num_classes, filters),
optimizer_fn=lambda x: torch.optim.Adam(params=x, lr=learning_rate),
model_name="unet3d_3plus")
network = fe.Network(ops=[
Resize3D(inputs="image", outputs="image", output_shape=input_shape),
Resize3D(inputs="label", outputs="label", output_shape=input_shape, mode='!infer'),
ModelOp(inputs="image", model=model, outputs="pred_segment"),
CrossEntropy(inputs=("pred_segment", "label"), outputs="ce_loss", form="binary"),
UpdateOp(model=model, loss_name="ce_loss"),
])
Training loop and Metrics¶
traces = [
Dice(
true_key="label",
pred_key="pred_segment",
channel_mapping={
0: 'Cell',
1: 'Mitochondria',
2: 'AlphaGranule',
3: 'CanalicularVessel',
4: 'GranuleBody',
5: 'GranuleCore'
}),
ReduceLROnPlateau(model=model, metric="Dice", patience=4, factor=0.5, best_mode="max"),
BestModelSaver(model=model, save_dir=save_dir, metric='Dice', save_best_mode='max'),
EarlyStopping(monitor="Dice", compare='max', min_delta=0.005, patience=6),
]
Let's start training¶
The training requires 40 epochs, and the total training time is around 28 mins hours on single Nvidia A100 (32G) GPU.
estimator = fe.Estimator(network=network,
pipeline=pipeline,
epochs=epochs,
log_steps=log_steps,
traces=traces,
train_steps_per_epoch=train_steps_per_epoch,
eval_steps_per_epoch=eval_steps_per_epoch)
estimator.fit()
______ __ ______ __ _ __
/ ____/___ ______/ /_/ ____/____/ /_(_)___ ___ ____ _/ /_____ _____
/ /_ / __ `/ ___/ __/ __/ / ___/ __/ / __ `__ \/ __ `/ __/ __ \/ ___/
/ __/ / /_/ (__ ) /_/ /___(__ ) /_/ / / / / / / /_/ / /_/ /_/ / /
/_/ \__,_/____/\__/_____/____/\__/_/_/ /_/ /_/\__,_/\__/\____/_/
FastEstimator-Start: step: 1; logging_interval: 20; num_device: 1;
FastEstimator-Train: step: 1; ce_loss: 0.8303619;
FastEstimator-Train: step: 20; ce_loss: 0.20598623; steps/sec: 1.36;
FastEstimator-Train: step: 40; ce_loss: 0.2167762; steps/sec: 1.3;
FastEstimator-Train: step: 60; ce_loss: 0.10972291; steps/sec: 1.29;
FastEstimator-Train: step: 75; epoch: 1; epoch_time(sec): 61.31;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.76;
Eval Progress: 16/25; steps/sec: 3.87;
Eval Progress: 25/25; steps/sec: 3.87;
FastEstimator-BestModelSaver: Saved model to /tmp/tmp79f0o1xf/unet3d_3plus_best_Dice.pt
FastEstimator-Eval: step: 75; epoch: 1; ce_loss: 0.1153822; Dice: 0.18591939; Dice_AlphaGranule: 0.0; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.87751913; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.22245294; Dice_Mitochondria: 0.015544222; max_Dice: 0.18591939; since_best_Dice: 0;
FastEstimator-Train: step: 80; ce_loss: 0.09550734; steps/sec: 1.09;
FastEstimator-Train: step: 100; ce_loss: 0.14471439; steps/sec: 1.29;
FastEstimator-Train: step: 120; ce_loss: 0.09462903; steps/sec: 1.29;
FastEstimator-Train: step: 140; ce_loss: 0.11916973; steps/sec: 1.29;
FastEstimator-Train: step: 150; epoch: 2; epoch_time(sec): 61.0;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 3.29;
Eval Progress: 16/25; steps/sec: 3.98;
Eval Progress: 25/25; steps/sec: 3.8;
FastEstimator-BestModelSaver: Saved model to /tmp/tmp79f0o1xf/unet3d_3plus_best_Dice.pt
FastEstimator-Eval: step: 150; epoch: 2; ce_loss: 0.09409454; Dice: 0.20101668; Dice_AlphaGranule: 0.051644072; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.9023661; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.25208992; Dice_Mitochondria: 0.0; max_Dice: 0.20101668; since_best_Dice: 0;
FastEstimator-Train: step: 160; ce_loss: 0.12400985; steps/sec: 1.08;
FastEstimator-Train: step: 180; ce_loss: 0.11078338; steps/sec: 1.29;
FastEstimator-Train: step: 200; ce_loss: 0.14163417; steps/sec: 1.29;
FastEstimator-Train: step: 220; ce_loss: 0.07257796; steps/sec: 1.29;
FastEstimator-Train: step: 225; epoch: 3; epoch_time(sec): 61.12;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.68;
Eval Progress: 16/25; steps/sec: 3.81;
Eval Progress: 25/25; steps/sec: 3.83;
FastEstimator-BestModelSaver: Saved model to /tmp/tmp79f0o1xf/unet3d_3plus_best_Dice.pt
FastEstimator-Eval: step: 225; epoch: 3; ce_loss: 0.10210272; Dice: 0.23396462; Dice_AlphaGranule: 0.15497324; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.8865098; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.3623047; Dice_Mitochondria: 0.0; max_Dice: 0.23396462; since_best_Dice: 0;
FastEstimator-Train: step: 240; ce_loss: 0.11134132; steps/sec: 1.08;
FastEstimator-Train: step: 260; ce_loss: 0.07451703; steps/sec: 1.29;
FastEstimator-Train: step: 280; ce_loss: 0.12084487; steps/sec: 1.29;
FastEstimator-Train: step: 300; ce_loss: 0.08398765; steps/sec: 1.29;
FastEstimator-Train: step: 300; epoch: 4; epoch_time(sec): 61.24;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.34;
Eval Progress: 16/25; steps/sec: 3.97;
Eval Progress: 25/25; steps/sec: 3.98;
FastEstimator-Eval: step: 300; epoch: 4; ce_loss: 0.09700322; Dice: 0.21093337; Dice_AlphaGranule: 0.12728098; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.8885958; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.20934826; Dice_Mitochondria: 0.040375184; max_Dice: 0.23396462; since_best_Dice: 1;
FastEstimator-Train: step: 320; ce_loss: 0.04077779; steps/sec: 1.06;
FastEstimator-Train: step: 340; ce_loss: 0.0628498; steps/sec: 1.29;
FastEstimator-Train: step: 360; ce_loss: 0.076646835; steps/sec: 1.29;
FastEstimator-Train: step: 375; epoch: 5; epoch_time(sec): 61.51;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.63;
Eval Progress: 16/25; steps/sec: 2.99;
Eval Progress: 25/25; steps/sec: 3.64;
FastEstimator-Eval: step: 375; epoch: 5; ce_loss: 0.124432154; Dice: 0.22382732; Dice_AlphaGranule: 0.059655525; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.8086924; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.46063298; Dice_Mitochondria: 0.013983042; max_Dice: 0.23396462; since_best_Dice: 2;
FastEstimator-Train: step: 380; ce_loss: 0.096815206; steps/sec: 1.07;
FastEstimator-Train: step: 400; ce_loss: 0.06408931; steps/sec: 1.29;
FastEstimator-Train: step: 420; ce_loss: 0.073252045; steps/sec: 1.29;
FastEstimator-Train: step: 440; ce_loss: 0.12816043; steps/sec: 1.29;
FastEstimator-Train: step: 450; epoch: 6; epoch_time(sec): 61.36;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 3.52;
Eval Progress: 16/25; steps/sec: 3.74;
Eval Progress: 25/25; steps/sec: 3.77;
FastEstimator-Eval: step: 450; epoch: 6; ce_loss: 0.10191353; Dice: 0.18513083; Dice_AlphaGranule: 0.17053589; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.8810749; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.059174225; Dice_Mitochondria: 0.0; max_Dice: 0.23396462; since_best_Dice: 3;
FastEstimator-Train: step: 460; ce_loss: 0.061374784; steps/sec: 1.09;
FastEstimator-Train: step: 480; ce_loss: 0.050663415; steps/sec: 1.29;
FastEstimator-Train: step: 500; ce_loss: 0.071846224; steps/sec: 1.29;
FastEstimator-Train: step: 520; ce_loss: 0.045135394; steps/sec: 1.29;
FastEstimator-Train: step: 525; epoch: 7; epoch_time(sec): 61.02;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.45;
Eval Progress: 16/25; steps/sec: 3.82;
Eval Progress: 25/25; steps/sec: 3.83;
FastEstimator-BestModelSaver: Saved model to /tmp/tmp79f0o1xf/unet3d_3plus_best_Dice.pt
FastEstimator-Eval: step: 525; epoch: 7; ce_loss: 0.08931035; Dice: 0.25831088; Dice_AlphaGranule: 0.20986092; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.89920825; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.43222672; Dice_Mitochondria: 0.008569288; max_Dice: 0.25831088; since_best_Dice: 0;
FastEstimator-Train: step: 540; ce_loss: 0.06356756; steps/sec: 1.06;
FastEstimator-Train: step: 560; ce_loss: 0.044075597; steps/sec: 1.29;
FastEstimator-Train: step: 580; ce_loss: 0.07535008; steps/sec: 1.29;
FastEstimator-Train: step: 600; ce_loss: 0.119257554; steps/sec: 1.29;
FastEstimator-Train: step: 600; epoch: 8; epoch_time(sec): 61.56;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.66;
Eval Progress: 16/25; steps/sec: 3.86;
Eval Progress: 25/25; steps/sec: 3.86;
FastEstimator-Eval: step: 600; epoch: 8; ce_loss: 0.09157195; Dice: 0.2567916; Dice_AlphaGranule: 0.22130911; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.89334536; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.42605677; Dice_Mitochondria: 3.8397422e-05; max_Dice: 0.25831088; since_best_Dice: 1;
FastEstimator-Train: step: 620; ce_loss: 0.11480411; steps/sec: 1.06;
FastEstimator-Train: step: 640; ce_loss: 0.065973036; steps/sec: 1.29;
FastEstimator-Train: step: 660; ce_loss: 0.043109555; steps/sec: 1.29;
FastEstimator-Train: step: 675; epoch: 9; epoch_time(sec): 61.51;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.11;
Eval Progress: 16/25; steps/sec: 3.76;
Eval Progress: 25/25; steps/sec: 3.67;
FastEstimator-BestModelSaver: Saved model to /tmp/tmp79f0o1xf/unet3d_3plus_best_Dice.pt
FastEstimator-Eval: step: 675; epoch: 9; ce_loss: 0.08640356; Dice: 0.27190855; Dice_AlphaGranule: 0.2839808; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.9077105; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.4354158; Dice_Mitochondria: 0.00434443; max_Dice: 0.27190855; since_best_Dice: 0;
FastEstimator-Train: step: 680; ce_loss: 0.0457928; steps/sec: 1.07;
FastEstimator-Train: step: 700; ce_loss: 0.050020874; steps/sec: 1.29;
FastEstimator-Train: step: 720; ce_loss: 0.065154016; steps/sec: 1.29;
FastEstimator-Train: step: 740; ce_loss: 0.042573478; steps/sec: 1.29;
FastEstimator-Train: step: 750; epoch: 10; epoch_time(sec): 61.28;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.79;
Eval Progress: 16/25; steps/sec: 3.48;
Eval Progress: 25/25; steps/sec: 3.64;
FastEstimator-Eval: step: 750; epoch: 10; ce_loss: 0.08486684; Dice: 0.24146147; Dice_AlphaGranule: 0.17277822; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.9102668; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.36550948; Dice_Mitochondria: 0.00021428218; max_Dice: 0.27190855; since_best_Dice: 1;
FastEstimator-Train: step: 760; ce_loss: 0.052132014; steps/sec: 1.06;
FastEstimator-Train: step: 780; ce_loss: 0.051481765; steps/sec: 1.29;
FastEstimator-Train: step: 800; ce_loss: 0.04546015; steps/sec: 1.29;
FastEstimator-Train: step: 820; ce_loss: 0.060806837; steps/sec: 1.29;
FastEstimator-Train: step: 825; epoch: 11; epoch_time(sec): 61.61;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.34;
Eval Progress: 16/25; steps/sec: 3.55;
Eval Progress: 25/25; steps/sec: 3.75;
FastEstimator-BestModelSaver: Saved model to /tmp/tmp79f0o1xf/unet3d_3plus_best_Dice.pt
FastEstimator-Eval: step: 825; epoch: 11; ce_loss: 0.07770658; Dice: 0.29375368; Dice_AlphaGranule: 0.35359073; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.91010475; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.49567142; Dice_Mitochondria: 0.0031552631; max_Dice: 0.29375368; since_best_Dice: 0;
FastEstimator-Train: step: 840; ce_loss: 0.04099419; steps/sec: 1.07;
FastEstimator-Train: step: 860; ce_loss: 0.03913365; steps/sec: 1.29;
FastEstimator-Train: step: 880; ce_loss: 0.05281667; steps/sec: 1.29;
FastEstimator-Train: step: 900; ce_loss: 0.042195227; steps/sec: 1.29;
FastEstimator-Train: step: 900; epoch: 12; epoch_time(sec): 61.45;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 3.13;
Eval Progress: 16/25; steps/sec: 3.04;
Eval Progress: 25/25; steps/sec: 3.4;
FastEstimator-Eval: step: 900; epoch: 12; ce_loss: 0.087085485; Dice: 0.2742488; Dice_AlphaGranule: 0.35462692; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.89047503; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.39421427; Dice_Mitochondria: 0.0061766254; max_Dice: 0.29375368; since_best_Dice: 1;
FastEstimator-Train: step: 920; ce_loss: 0.09527723; steps/sec: 1.07;
FastEstimator-Train: step: 940; ce_loss: 0.048805945; steps/sec: 1.29;
FastEstimator-Train: step: 960; ce_loss: 0.064195104; steps/sec: 1.29;
FastEstimator-Train: step: 975; epoch: 13; epoch_time(sec): 61.36;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 3.1;
Eval Progress: 16/25; steps/sec: 3.51;
Eval Progress: 25/25; steps/sec: 3.87;
FastEstimator-Eval: step: 975; epoch: 13; ce_loss: 0.0977592; Dice: 0.21654314; Dice_AlphaGranule: 0.365906; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.89077944; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.04257338; Dice_Mitochondria: 0.0; max_Dice: 0.29375368; since_best_Dice: 2;
FastEstimator-Train: step: 980; ce_loss: 0.057625346; steps/sec: 1.08;
FastEstimator-Train: step: 1000; ce_loss: 0.06405907; steps/sec: 1.29;
FastEstimator-Train: step: 1020; ce_loss: 0.041967522; steps/sec: 1.29;
FastEstimator-Train: step: 1040; ce_loss: 0.054536086; steps/sec: 1.29;
FastEstimator-Train: step: 1050; epoch: 14; epoch_time(sec): 61.26;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.57;
Eval Progress: 16/25; steps/sec: 3.87;
Eval Progress: 25/25; steps/sec: 3.74;
FastEstimator-Eval: step: 1050; epoch: 14; ce_loss: 0.10027878; Dice: 0.2677289; Dice_AlphaGranule: 0.3271591; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.8415446; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.43572918; Dice_Mitochondria: 0.001940464; max_Dice: 0.29375368; since_best_Dice: 3;
FastEstimator-Train: step: 1060; ce_loss: 0.054968704; steps/sec: 1.06;
FastEstimator-Train: step: 1080; ce_loss: 0.036659826; steps/sec: 1.29;
FastEstimator-Train: step: 1100; ce_loss: 0.035622057; steps/sec: 1.29;
FastEstimator-Train: step: 1120; ce_loss: 0.026277982; steps/sec: 1.29;
FastEstimator-Train: step: 1125; epoch: 15; epoch_time(sec): 61.56;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.89;
Eval Progress: 16/25; steps/sec: 3.43;
Eval Progress: 25/25; steps/sec: 3.46;
FastEstimator-ReduceLROnPlateau: learning rate reduced to 0.0005000000237487257
FastEstimator-Eval: step: 1125; epoch: 15; ce_loss: 0.090255305; Dice: 0.2352698; Dice_AlphaGranule: 0.2562945; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.9112644; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.24402592; Dice_Mitochondria: 3.398266e-05; max_Dice: 0.29375368; since_best_Dice: 4; unet3d_3plus_lr: 0.0005;
FastEstimator-Train: step: 1140; ce_loss: 0.045861237; steps/sec: 1.08;
FastEstimator-Train: step: 1160; ce_loss: 0.06833782; steps/sec: 1.29;
FastEstimator-Train: step: 1180; ce_loss: 0.022313815; steps/sec: 1.29;
FastEstimator-Train: step: 1200; ce_loss: 0.044976972; steps/sec: 1.29;
FastEstimator-Train: step: 1200; epoch: 16; epoch_time(sec): 61.19;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.04;
Eval Progress: 16/25; steps/sec: 3.85;
Eval Progress: 25/25; steps/sec: 3.97;
FastEstimator-Eval: step: 1200; epoch: 16; ce_loss: 0.08719411; Dice: 0.25694326; Dice_AlphaGranule: 0.30967927; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.908896; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.32086614; Dice_Mitochondria: 0.002218212; max_Dice: 0.29375368; since_best_Dice: 5;
FastEstimator-Train: step: 1220; ce_loss: 0.05291032; steps/sec: 1.08;
FastEstimator-Train: step: 1240; ce_loss: 0.07368225; steps/sec: 1.29;
FastEstimator-Train: step: 1260; ce_loss: 0.035692345; steps/sec: 1.29;
FastEstimator-Train: step: 1275; epoch: 17; epoch_time(sec): 61.29;
Eval Progress: 1/25;
Eval Progress: 8/25; steps/sec: 2.01;
Eval Progress: 16/25; steps/sec: 3.84;
Eval Progress: 25/25; steps/sec: 3.02;
FastEstimator-EarlyStopping: 'Dice' triggered an early stop. Its best value was 0.2937536835670471 at epoch 11
FastEstimator-Eval: step: 1275; epoch: 17; ce_loss: 0.08027802; Dice: 0.2673981; Dice_AlphaGranule: 0.34898543; Dice_CanalicularVessel: 0.0; Dice_Cell: 0.9117461; Dice_GranuleBody: 0.0; Dice_GranuleCore: 0.34234184; Dice_Mitochondria: 0.0013150654; max_Dice: 0.29375368; since_best_Dice: 6;
FastEstimator-Finish: step: 1275; total_time(sec): 1228.73; unet3d_3plus_lr: 0.0005;
Inferencing¶
After training the network, let's inference our trained model and visualize their results in comparison to the ground truth. For visualization, we will use validation data.
sample_val_batch = pipeline.get_results(mode="eval", shuffle=True)
sample_val_batch = network.transform(data=sample_val_batch, mode="eval")
Visualizing sample segmentation prediction¶
image = sample_val_batch['image'].numpy()
label = sample_val_batch['label'].numpy()
print("Image shape: ", image.shape," Mask shape:", label.shape)
image = np.squeeze(image[0])
label = np.transpose(label[0], (1, 2, 3, 0))
pred_segment = np.transpose(sample_val_batch['pred_segment'].numpy()[0], (1, 2, 3, 0))
pred_segment[pred_segment>0.5]=1
pred_segment[pred_segment<0.5]=0
Image shape: (1, 1, 256, 256, 24) Mask shape: (1, 6, 256, 256, 24)
Now lets visualize inference output, a single slice of image with the label overlaying on it using ImageDisplay.
GridDisplay([ImageDisplay(image=image[:,:,0], title='Image'),
ImageDisplay(image=image[:,:,0], masks=label[:,:,0], title='Ground Truth'),
ImageDisplay(image=image[:,:,0], masks=pred_segment[:,:,0], title='Prediction')
]).show()
As you can see, while the model performance is not great on classes like cancalicular vessel, granule body and mitochondria because of class imbalance, since they constitute only small fraction of the image. The model is doing a great job in segmenting Cell,and decent performance on alpha granule and granule core. Overall UNET 3D plus is a efficient model for 3D segmentation.
If you want you visualize the all the slices of inference output overlayed on the image, please use below code to save multi slice visualization as gif.
# adding ground truth layer
z = np.ones(pred_segment.shape[:-1] + (1,), dtype=pred_segment.dtype)
z[np.sum(pred_segment, axis=-1)==1] = 0
pred_segment_out = np.concatenate((z, pred_segment), axis=-1)
image = np.tile(np.expand_dims(image, -1), 3)
save_gif(image, pred_segment_out, color_mapping, 'prediction.gif')
A sample of the visualization of multiple slices of the inference output which is generated offline.
