Preliminary Setup¶
We will first set up a basic MNIST example for the rest of the demonstrations:
import fastestimator as fe
from fastestimator.architecture.tensorflow import LeNet
from fastestimator.dataset.data import mnist
from fastestimator.op.numpyop.univariate import ExpandDims, Minmax
from fastestimator.op.tensorop.loss import CrossEntropy
from fastestimator.op.tensorop.model import ModelOp, UpdateOp
from fastestimator.schedule import cosine_decay
from fastestimator.trace.adapt import LRScheduler
from fastestimator.trace.metric import Accuracy
from fastestimator.trace.io import TensorBoard
train_data, eval_data = mnist.load_data()
test_data = eval_data.split(0.5)
pipeline = fe.Pipeline(train_data=train_data,
eval_data=eval_data,
test_data=test_data,
batch_size=32,
ops=[ExpandDims(inputs="x", outputs="x"), Minmax(inputs="x", outputs="x")])
model = fe.build(model_fn=LeNet, optimizer_fn="adam")
network = fe.Network(ops=[
ModelOp(model=model, inputs="x", outputs="y_pred"),
CrossEntropy(inputs=("y_pred", "y"), outputs="ce"),
UpdateOp(model=model, loss_name="ce")
])
traces = [
Accuracy(true_key="y", pred_key="y_pred"),
LRScheduler(model=model, lr_fn=lambda step: cosine_decay(step, cycle_length=3750, init_lr=1e-3))
]
Experiment Logging¶
As you may have noticed if you have used FastEstimator, log messages are printed to the screen during training. If you want to persist these log messages for later records, you can simply pipe them into a file when launching training from the command line, or else just copy and paste the messages from the console into a persistent file on the disk. FastEstimator allows logging to be controlled via arguments passed to the Estimator class, as described in the Beginner Tutorial 7. Let's see an example logging every 120 steps:
est = fe.Estimator(pipeline=pipeline, network=network, epochs=1, traces=traces, log_steps=120)
est.fit()
______ __ ______ __ _ __
/ ____/___ ______/ /_/ ____/____/ /_(_)___ ___ ____ _/ /_____ _____
/ /_ / __ `/ ___/ __/ __/ / ___/ __/ / __ `__ \/ __ `/ __/ __ \/ ___/
/ __/ / /_/ (__ ) /_/ /___(__ ) /_/ / / / / / / /_/ / /_/ /_/ / /
/_/ \__,_/____/\__/_____/____/\__/_/_/ /_/ /_/\__,_/\__/\____/_/
FastEstimator-Warn: No ModelSaver Trace detected. Models will not be saved.
FastEstimator-Start: step: 1; logging_interval: 120; num_device: 0;
FastEstimator-Train: step: 1; ce: 2.2963042; model_lr: 0.0009999998;
FastEstimator-Train: step: 120; ce: 0.1863498; model_lr: 0.000997478; steps/sec: 70.22;
FastEstimator-Train: step: 240; ce: 0.051570907; model_lr: 0.0009899376; steps/sec: 70.85;
FastEstimator-Train: step: 360; ce: 0.14517793; model_lr: 0.0009774548; steps/sec: 69.82;
FastEstimator-Train: step: 480; ce: 0.16006204; model_lr: 0.0009601558; steps/sec: 71.39;
FastEstimator-Train: step: 600; ce: 0.014987067; model_lr: 0.0009382152; steps/sec: 70.44;
FastEstimator-Train: step: 720; ce: 0.14745927; model_lr: 0.00091185456; steps/sec: 67.46;
FastEstimator-Train: step: 840; ce: 0.06190053; model_lr: 0.00088134; steps/sec: 69.75;
FastEstimator-Train: step: 960; ce: 0.0073453495; model_lr: 0.00084697985; steps/sec: 68.31;
FastEstimator-Train: step: 1080; ce: 0.0628736; model_lr: 0.0008091209; steps/sec: 68.8;
FastEstimator-Train: step: 1200; ce: 0.09909142; model_lr: 0.0007681455; steps/sec: 68.23;
FastEstimator-Train: step: 1320; ce: 0.006691601; model_lr: 0.0007244674; steps/sec: 68.03;
FastEstimator-Train: step: 1440; ce: 0.031444274; model_lr: 0.00067852775; steps/sec: 64.0;
FastEstimator-Train: step: 1560; ce: 0.0026731049; model_lr: 0.0006307903; steps/sec: 66.83;
FastEstimator-Train: step: 1680; ce: 0.2757488; model_lr: 0.00058173726; steps/sec: 61.66;
FastEstimator-Train: step: 1800; ce: 0.1322045; model_lr: 0.0005318639; steps/sec: 56.45;
FastEstimator-Train: step: 1875; epoch: 1; epoch_time: 29.41 sec;
FastEstimator-Eval: step: 1875; epoch: 1; accuracy: 0.9876; ce: 0.035917815;
FastEstimator-Finish: step: 1875; model_lr: 0.0005005; total_time: 30.63 sec;
Experiment Summaries¶
Having log messages on the screen can be handy, but what if you want to access these messages within python? Enter the Summary class. Summary objects contain information about the training over time, and will be automatically generated when the Estimator fit() method is invoked with an experiment name:
est = fe.Estimator(pipeline=pipeline, network=network, epochs=1, traces=traces, log_steps=500)
summary = est.fit("experiment1")
______ __ ______ __ _ __
/ ____/___ ______/ /_/ ____/____/ /_(_)___ ___ ____ _/ /_____ _____
/ /_ / __ `/ ___/ __/ __/ / ___/ __/ / __ `__ \/ __ `/ __/ __ \/ ___/
/ __/ / /_/ (__ ) /_/ /___(__ ) /_/ / / / / / / /_/ / /_/ /_/ / /
/_/ \__,_/____/\__/_____/____/\__/_/_/ /_/ /_/\__,_/\__/\____/_/
FastEstimator-Warn: No ModelSaver Trace detected. Models will not be saved.
FastEstimator-Start: step: 1; logging_interval: 500; num_device: 0;
FastEstimator-Train: step: 1; ce: 0.12709273; model_lr: 0.0009999998;
FastEstimator-Train: step: 500; ce: 0.0039903466; model_lr: 0.00095681596; steps/sec: 63.74;
FastEstimator-Train: step: 1000; ce: 0.1970906; model_lr: 0.00083473074; steps/sec: 66.85;
FastEstimator-Train: step: 1500; ce: 0.00413731; model_lr: 0.000654854; steps/sec: 66.22;
FastEstimator-Train: step: 1875; epoch: 1; epoch_time: 29.32 sec;
FastEstimator-Eval: step: 1875; epoch: 1; accuracy: 0.988; ce: 0.03613729;
FastEstimator-Finish: step: 1875; model_lr: 0.0005005; total_time: 30.65 sec;
Lets take a look at what sort of information is contained within our Summary object:
summary.name
'experiment1'
summary.history
defaultdict(<function fastestimator.summary.summary.Summary.__init__.<locals>.<lambda>()>,
{'train': defaultdict(dict,
{'logging_interval': {0: array(500)},
'num_device': {0: array(0)},
'ce': {1: array(0.12709273, dtype=float32),
500: array(0.00399035, dtype=float32),
1000: array(0.1970906, dtype=float32),
1500: array(0.00413731, dtype=float32)},
'model_lr': {1: array(0.001, dtype=float32),
500: array(0.00095682, dtype=float32),
1000: array(0.00083473, dtype=float32),
1500: array(0.00065485, dtype=float32),
1875: array(0.0005005, dtype=float32)},
'steps/sec': {500: array(63.74),
1000: array(66.85),
1500: array(66.22)},
'epoch': {1875: 1},
'epoch_time': {1875: array('29.32 sec', dtype='<U9')},
'total_time': {1875: array('30.65 sec', dtype='<U9')}}),
'eval': defaultdict(dict,
{'epoch': {1875: 1},
'accuracy': {1875: array(0.988)},
'ce': {1875: array(0.03613729, dtype=float32)}})})
The history field can appear a little daunting, but it is simply a dictionary laid out as follows: {mode: {key: {step: value}}}. Once you have invoked the .fit() method with an experiment name, subsequent calls to .test() will add their results into the same summary dictionary:
summary = est.test()
FastEstimator-Test: step: 1875; epoch: 1; accuracy: 0.9896; ce: 0.03368678;
summary.history
defaultdict(<function fastestimator.summary.summary.Summary.__init__.<locals>.<lambda>()>,
{'train': defaultdict(dict,
{'logging_interval': {0: array(500)},
'num_device': {0: array(0)},
'ce': {1: array(0.12709273, dtype=float32),
500: array(0.00399035, dtype=float32),
1000: array(0.1970906, dtype=float32),
1500: array(0.00413731, dtype=float32)},
'model_lr': {1: array(0.001, dtype=float32),
500: array(0.00095682, dtype=float32),
1000: array(0.00083473, dtype=float32),
1500: array(0.00065485, dtype=float32),
1875: array(0.0005005, dtype=float32)},
'steps/sec': {500: array(63.74),
1000: array(66.85),
1500: array(66.22)},
'epoch': {1875: 1},
'epoch_time': {1875: array('29.32 sec', dtype='<U9')},
'total_time': {1875: array('30.65 sec', dtype='<U9')}}),
'eval': defaultdict(dict,
{'epoch': {1875: 1},
'accuracy': {1875: array(0.988)},
'ce': {1875: array(0.03613729, dtype=float32)}}),
'test': defaultdict(dict,
{'epoch': {1875: 1},
'accuracy': {1875: array(0.9896)},
'ce': {1875: array(0.03368678, dtype=float32)}})})
Even if an experiment name was not provided during the .fit() call, it may be provided during the .test() call. The resulting summary object will, however, only contain information from the Test mode.
Log Parsing¶
Suppose that you have a log file saved to disk, and you want to create an in-memory Summary representation of it. This can be done through FastEstimator logging utilities:
summary = fe.summary.logs.parse_log_file(file_path="../resources/t06a_exp1.txt", file_extension=".txt")
summary.name
't06a_exp1'
summary.history['eval']
defaultdict(dict,
{'epoch': {1875: 1.0, 3750: 2.0, 5625: 3.0},
'ce': {1875: 0.03284014, 3750: 0.02343675, 5625: 0.02382297},
'min_ce': {1875: 0.03284014, 3750: 0.02343675, 5625: 0.02343675},
'since_best': {1875: 0.0, 3750: 0.0, 5625: 1.0},
'accuracy': {1875: 0.9882, 3750: 0.992, 5625: 0.9922}})
Log Visualization¶
While seeing log data as numbers can be informative, visualizations of data are often more useful. FastEstimator provides several ways to visualize log data: from python using Summary objects or log files, as well as through the command line.
fe.summary.logs.visualize_logs(experiments=[summary])
If you are only interested in visualizing a subset of these log values, it is also possible to whitelist or blacklist values via the 'include_metrics' and 'ignore_metrics' arguments respectively:
fe.summary.logs.visualize_logs(experiments=[summary], include_metrics={"accuracy", "ce"})
It is also possible to compare logs from different experiments, which can be especially useful when fiddling with hyper-parameter values to determine their effects on training:
fe.summary.logs.parse_log_files(file_paths=["../resources/t06a_exp1.txt", "../resources/t06a_exp2.txt"], log_extension=".txt")
All of the log files within a given directory can also be compared at the same time, either by using the parse_log_dir() method or via the command line as follows: fastestimator logs --extension .txt --smooth 0 ../resources
Visualizing Repeat Trials¶
Suppose you are running some experiments like the ones above to try and decide which of several experimental configurations is best. For example, suppose you are trying to decide between lossA and lossB. You run 5 experiments with each loss in order to account for randomness, and save the logs as lossA_1.txt, lossA_2.txt, lossB_1.txt, etc. You could use the method described above, for example:
fe.summary.logs.parse_log_dir(dir_path='../resources/t06a_logs', smooth_factor=0)
While this is certainly an option, it is not very easy to tell at a glance which of lossA or lossB is superior. Let's use log grouping in order to get a cleaner picture:
fe.summary.logs.parse_log_dir(dir_path='../resources/t06a_logs', smooth_factor=0, group_by=r'(.*)_[\d]+\.txt')
Now we are displaying the mean values for lossA and lossB, plus or minus their standard deviations over the 5 experiments. This makes it easy to see that lossA results in a better mcc score and calibration error, whereas lossB has slightly faster training, but the speeds are typically within 1 standard deviation so that might be noise. The group_by argument can take any regex pattern, and if you are using it from the command line, you can simply pass --group_by _n as a shortcut to get the regex pattern used above.
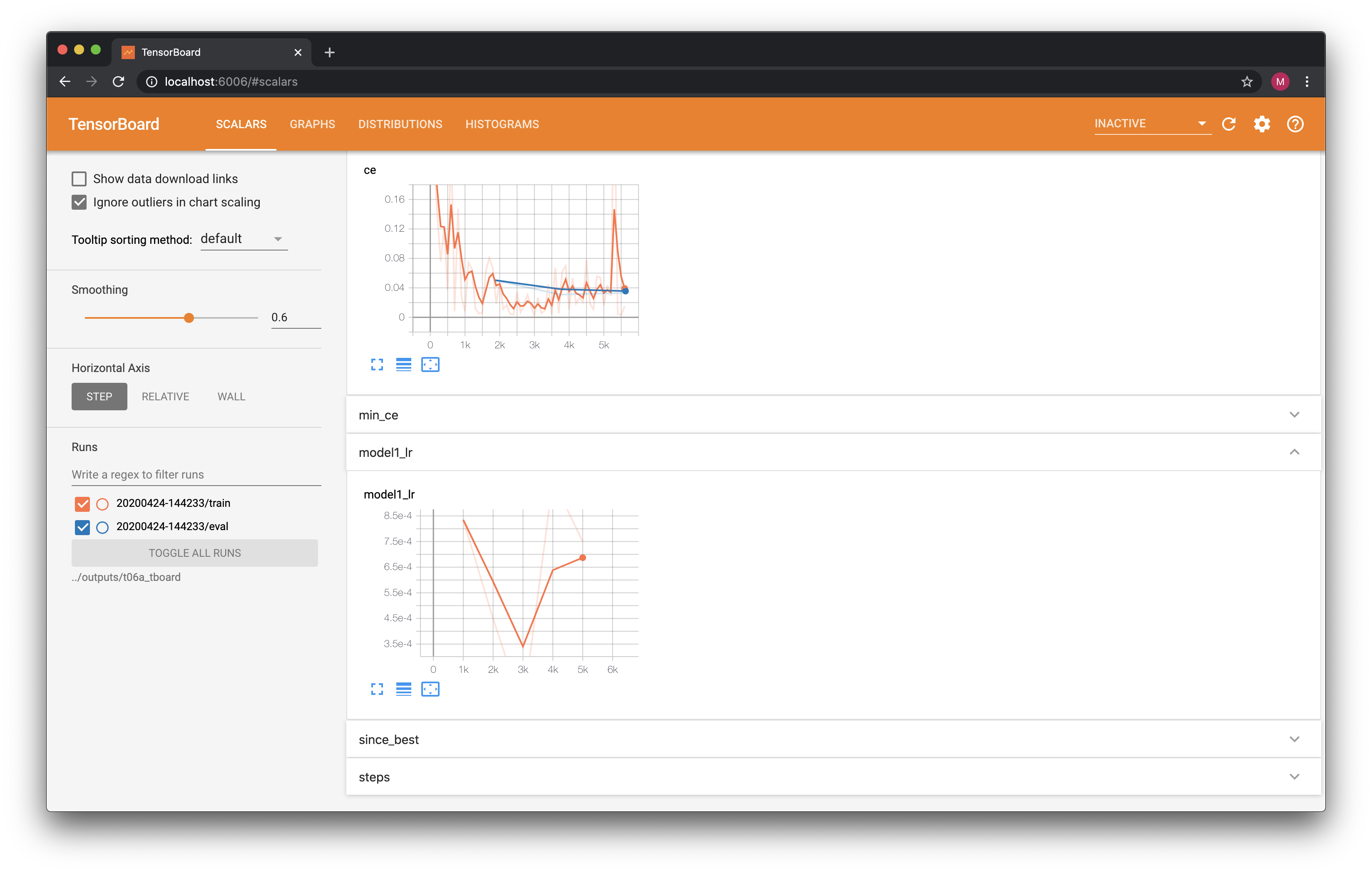
TensorBoard¶
Of course, no modern AI framework would be complete without TensorBoard integration. In FastEstimator, all that is required to achieve TensorBoard integration is to add the TensorBoard Trace to the list of traces passed to the Estimator:
import tempfile
log_dir = tempfile.mkdtemp()
pipeline = fe.Pipeline(train_data=train_data,
eval_data=eval_data,
test_data=test_data,
batch_size=32,
ops=[ExpandDims(inputs="x", outputs="x"), Minmax(inputs="x", outputs="x")], num_process=0)
model = fe.build(model_fn=LeNet, optimizer_fn="adam")
network = fe.Network(ops=[
ModelOp(model=model, inputs="x", outputs="y_pred"),
CrossEntropy(inputs=("y_pred", "y"), outputs="ce"),
UpdateOp(model=model, loss_name="ce")
])
traces = [
Accuracy(true_key="y", pred_key="y_pred"),
LRScheduler(model=model, lr_fn=lambda step: cosine_decay(step, cycle_length=3750, init_lr=1e-3)),
TensorBoard(log_dir=log_dir, weight_histogram_freq="epoch")
]
est = fe.Estimator(pipeline=pipeline, network=network, epochs=3, traces=traces, log_steps=1000)
est.fit()
______ __ ______ __ _ __
/ ____/___ ______/ /_/ ____/____/ /_(_)___ ___ ____ _/ /_____ _____
/ /_ / __ `/ ___/ __/ __/ / ___/ __/ / __ `__ \/ __ `/ __/ __ \/ ___/
/ __/ / /_/ (__ ) /_/ /___(__ ) /_/ / / / / / / /_/ / /_/ /_/ / /
/_/ \__,_/____/\__/_____/____/\__/_/_/ /_/ /_/\__,_/\__/\____/_/
FastEstimator-Warn: No ModelSaver Trace detected. Models will not be saved.
FastEstimator-Tensorboard: writing logs to /var/folders/3r/h9kh47050gv6rbt_pgf8cl540000gn/T/tmpnb971yiz/20220413-150813
FastEstimator-Start: step: 1; logging_interval: 1000; num_device: 0;
WARNING:tensorflow:5 out of the last 160 calls to <function TFNetwork._forward_step_static at 0x1814cf790> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
FastEstimator-Train: step: 1; ce: 2.3051662; model1_lr: 0.0009999998;
FastEstimator-Train: step: 1000; ce: 0.3109427; model1_lr: 0.00083473074; steps/sec: 57.51;
FastEstimator-Train: step: 1875; epoch: 1; epoch_time: 33.49 sec;
FastEstimator-Eval: step: 1875; epoch: 1; accuracy: 0.9888; ce: 0.037164193;
FastEstimator-Train: step: 2000; ce: 0.0102050435; model1_lr: 0.00044828805; steps/sec: 53.36;
FastEstimator-Train: step: 3000; ce: 0.013866902; model1_lr: 9.639601e-05; steps/sec: 47.11;
FastEstimator-Train: step: 3750; epoch: 2; epoch_time: 40.77 sec;
FastEstimator-Eval: step: 3750; epoch: 2; accuracy: 0.9914; ce: 0.026301745;
FastEstimator-Train: step: 4000; ce: 0.010989559; model1_lr: 0.0009890847; steps/sec: 45.03;
FastEstimator-Train: step: 5000; ce: 0.007876373; model1_lr: 0.00075025; steps/sec: 45.57;
FastEstimator-Train: step: 5625; epoch: 3; epoch_time: 41.5 sec;
FastEstimator-Eval: step: 5625; epoch: 3; accuracy: 0.9888; ce: 0.034312796;
FastEstimator-Finish: step: 5625; model1_lr: 0.0005005; total_time: 121.21 sec;
Now let's launch TensorBoard to visualize our logs. Note that this call will prevent any subsequent Jupyter Notebook cells from running until you manually terminate it.
#!tensorboard --reload_multifile=true --logdir /var/folders/lx/drkxftt117gblvgsp1p39rlc0000gn/T/tmpb_oy2ihe
The TensorBoard display should look something like this: